SmootherExps
The SmootherExps.jl sub-module contains methods to configure filter twin experiments, using a stored time series as generated by GenerateTimeSeries as the underlying observation generating process. The frequency of observations in continuous time is defined by the frequency of data saved in the time series and is inferred by the experiment when reading in the data.
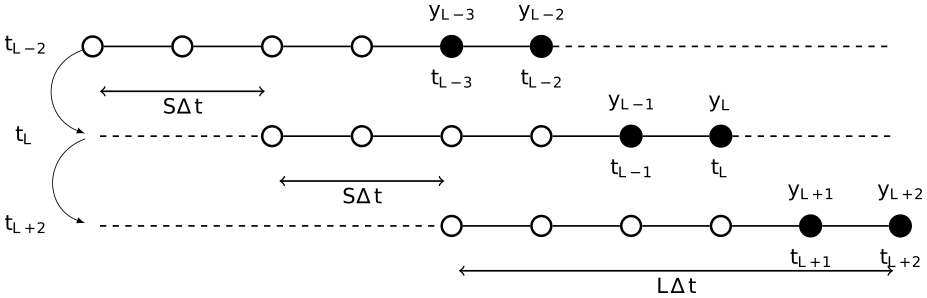
Smoother experiment configurations are generated by supplying a NamedTuple with the required fields as specified in the experiment method. Conventions for these arguments are the same as with the FilterExps, with the additional options that configure the data assimilation window (DAW) and how this is shifted in time:
lag- the number of past observation / analysis times to reanalyze in a DAW, corresponding to $L$ in the figure above;shift- the number of observation / analysis times to shift the DAW, corresponding to $S$ in the figure above;mda- (Multiple Data Assimilation), typeBool, determines whether the technique of multiple data assimilation is used (only compatible withsingle_iterationanditerativesmoothers.
Currently debugged and validated smoother experiment configurations include
classic_state- classic ETKS style state estimationclassic_param- classic ETKS style state-parameter estimationsingle_iteration_state- SIEnKS state estimationsingle_iteration_param- SIEnKS state-parameter estimationiterative_state- IEnKS Gauss-Newton style state estimationiterative_param- IEnKS Gauss-Newton style state-parameter estimation
Note, the single-iteration and fully iterative Gauss-Newton style smoothers are only defined for MDA compatible values of lag and shift where the lag is an integer multiple of the shift.
Smoother Experiment Methods
DataAssimilationBenchmarks.SmootherExps.classic_ensemble_param — Methodclassic_ensemble_param((time_series::String, method::String, seed::Int64, nanl::Int64,
lag::Int64, shift::Int64, obs_un::Float64, obs_dim::Int64,
γ::Float64, p_err::Float64, p_wlk::Float64, N_ens::Int64,
s_infl::Float64, s_infl::Float64})::NamedTuple)Classic ensemble Kalman smoother joint state-parameter estimation twin experiment.
NOTE: the classic scheme does not use multiple data assimilation and we hard code mda=false in the function for consistency with other methods.
Output from the experiment is saved in a dictionary of the form,
data = Dict{String,Any}(
"fore_rmse" => fore_rmse,
"filt_rmse" => filt_rmse,
"post_rmse" => post_rmse,
"param_rmse" => para_rmse,
"fore_spread" => fore_spread,
"filt_spread" => filt_spread,
"post_spread" => post_spread,
"param_spread" => para_spread,
"method" => method,
"seed" => seed,
"diffusion" => diffusion,
"dx_params" => dx_params,
"param_truth" => param_truth,
"sys_dim" => sys_dim,
"state_dim" => state_dim,
"obs_dim" => obs_dim,
"obs_un" => obs_un,
"γ" => γ,
"p_err" => p_err,
"p_wlk" => p_wlk,
"nanl" => nanl,
"tanl" => tanl,
"lag" => lag,
"shift" => shift,
"mda" => mda,
"h" => h,
"N_ens" => N_ens,
"s_infl" => round(s_infl, digits=2),
"p_infl" => round(p_infl, digits=2)
)
if haskey(ts, "diff_mat")
data["diff_mat"] = ts["diff_mat"]
endExperiment output is written to a directory defined by
path = pkgdir(DataAssimilationBenchmarks) * "/src/data/" * method * "-classic/"where the file name is written dynamically according to the selected parameters as follows:
method * "-classic_" * model *
"_state_seed_" * lpad(seed, 4, "0") *
"_diff_" * rpad(diffusion, 5, "0") *
"_sysD_" * lpad(sys_dim, 2, "0") *
"_obsD_" * lpad(obs_dim, 2, "0") *
"_obsU_" * rpad(obs_un, 4, "0") *
"_gamma_" * lpad(γ, 5, "0") *
"_nanl_" * lpad(nanl, 5, "0") *
"_tanl_" * rpad(tanl, 4, "0") *
"_h_" * rpad(h, 4, "0") *
"_lag_" * lpad(lag, 3, "0") *
"_shift_" * lpad(shift, 3, "0") *
"_mda_" * string(mda) *
"_nens_" * lpad(N_ens, 3,"0") *
"_stateInfl_" * rpad(round(s_infl, digits=2), 4, "0") *
".jld2"DataAssimilationBenchmarks.SmootherExps.classic_ensemble_state — Methodclassic_ensemble_state((time_series::String, method::String, seed::Int64, nanl::Int64,
lag::Int64, shift::Int64, obs_un::Float64, obs_dim::Int64,
γ::Float64, N_ens::Int64, s_infl::Float64)::NamedTuple)Classic ensemble Kalman smoother state estimation twin experiment.
NOTE: the classic scheme does not use multiple data assimilation and we hard code mda=false in the function for consistency with the API of other methods.
Output from the experiment is saved in a dictionary of the form,
data = Dict{String,Any}(
"fore_rmse" => fore_rmse,
"filt_rmse" => filt_rmse,
"post_rmse" => post_rmse,
"fore_spread" => fore_spread,
"filt_spread" => filt_spread,
"post_spread" => post_spread,
"method" => method,
"seed" => seed,
"diffusion" => diffusion,
"dx_params" => dx_params,
"sys_dim" => sys_dim,
"obs_dim" => obs_dim,
"obs_un" => obs_un,
"γ" => γ,
"nanl" => nanl,
"tanl" => tanl,
"lag" => lag,
"shift" => shift,
"h" => h,
"N_ens" => N_ens,
"mda" => mda,
"s_infl" => round(s_infl, digits=2)
)
if haskey(ts, "diff_mat")
data["diff_mat"] = ts["diff_mat"]
endExperiment output is written to a directory defined by
path = pkgdir(DataAssimilationBenchmarks) * "/src/data/" * method * "-classic/"where the file name is written dynamically according to the selected parameters as follows:
method * "-classic_" * model *
"_state_seed_" * lpad(seed, 4, "0") *
"_diff_" * rpad(diffusion, 5, "0") *
"_sysD_" * lpad(sys_dim, 2, "0") *
"_obsD_" * lpad(obs_dim, 2, "0") *
"_obsU_" * rpad(obs_un, 4, "0") *
"_gamma_" * lpad(γ, 5, "0") *
"_nanl_" * lpad(nanl, 5, "0") *
"_tanl_" * rpad(tanl, 4, "0") *
"_h_" * rpad(h, 4, "0") *
"_lag_" * lpad(lag, 3, "0") *
"_shift_" * lpad(shift, 3, "0") *
"_mda_" * string(mda) *
"_nens_" * lpad(N_ens, 3,"0") *
"_stateInfl_" * rpad(round(s_infl, digits=2), 4, "0") *
".jld2"DataAssimilationBenchmarks.SmootherExps.iterative_ensemble_param — Methoditerative_ensemble_param((time_series:String, method:String, seed::Int64, nanl::Int64,
lag::Int64, shift::Int64, mda::Bool, obs_un::Float64,
obs_dim::Int64, γ::Float64, p_err::Float64, p_wlk::Float64,
N_ens::Int64, s_infl::Float64, p_infl::Float64)::NamedTuple)4DEnVAR joint state-parameter estimation twin experiment using the IEnKS formalism.
Output from the experiment is saved in a dictionary of the form,
data = Dict{String,Any}(
"fore_rmse" => fore_rmse,
"filt_rmse" => filt_rmse,
"post_rmse" => post_rmse,
"param_rmse" => para_rmse,
"fore_spread" => fore_spread,
"filt_spread" => filt_spread,
"post_spread" => post_spread,
"param_spread" => para_spread,
"iteration_sequence" => iteration_sequence,
"method" => method,
"seed" => seed,
"diffusion" => diffusion,
"dx_params" => dx_params,
"sys_dim" => sys_dim,
"obs_dim" => obs_dim,
"obs_un" => obs_un,
"γ" => γ,
"p_wlk" => p_wlk,
"p_infl" => p_infl,
"nanl" => nanl,
"tanl" => tanl,
"lag" => lag,
"shift" => shift,
"mda" => mda,
"h" => h,
"N_ens" => N_ens,
"s_infl" => round(s_infl, digits=2),
"p_infl" => round(p_infl, digits=2)
)
if haskey(ts, "diff_mat")
data["diff_mat"] = ts["diff_mat"]
endExperiment output is written to a directory defined by
path = pkgdir(DataAssimilationBenchmarks) * "/src/data/" * method * "/"where the file name is written dynamically according to the selected parameters as follows:
method * "_" * model *
"_param_seed_" * lpad(seed, 4, "0") *
"_diff_" * rpad(diffusion, 5, "0") *
"_sysD_" * lpad(sys_dim, 2, "0") *
"_obsD_" * lpad(obs_dim, 2, "0") *
"_obsU_" * rpad(obs_un, 4, "0") *
"_gamma_" * lpad(γ, 5, "0") *
"_paramE_" * rpad(p_err, 4, "0") *
"_paramW_" * rpad(p_wlk, 6, "0") *
"_nanl_" * lpad(nanl, 5, "0") *
"_tanl_" * rpad(tanl, 4, "0") *
"_h_" * rpad(h, 4, "0") *
"_lag_" * lpad(lag, 3, "0") *
"_shift_" * lpad(shift, 3, "0") *
"_mda_" * string(mda) *
"_nens_" * lpad(N_ens, 3,"0") *
"_stateInfl_" * rpad(round(s_infl, digits=2), 4, "0") *
"_paramInfl_" * rpad(round(p_infl, digits=2), 4, "0") *
".jld2"DataAssimilationBenchmarks.SmootherExps.iterative_ensemble_state — Methoditerative_ensemble_state((time_series::String, method::String, seed::Int64, nanl::Int64,
lag::Int64, shift::Int64, mda::Bool, obs_un::Float64,
obs_dim::Int64, γ::Float64, N_ens::Int64,
s_infl::Float64)::NamedTuple)4DEnVAR state estimation twin experiment using the IEnKS formalism.
Output from the experiment is saved in a dictionary of the form,
data = Dict{String,Any}(
"fore_rmse" => fore_rmse,
"filt_rmse" => filt_rmse,
"post_rmse" => post_rmse,
"fore_spread" => fore_spread,
"filt_spread" => filt_spread,
"post_spread" => post_spread,
"iteration_sequence" => iteration_sequence,
"method" => method,
"seed" => seed,
"diffusion" => diffusion,
"dx_params" => dx_params,
"sys_dim" => sys_dim,
"obs_dim" => obs_dim,
"obs_un" => obs_un,
"γ" => γ,
"nanl" => nanl,
"tanl" => tanl,
"lag" => lag,
"shift" => shift,
"mda" => mda,
"h" => h,
"N_ens" => N_ens,
"s_infl" => round(s_infl, digits=2)
)
if haskey(ts, "diff_mat")
data["diff_mat"] = ts["diff_mat"]
endExperiment output is written to a directory defined by
path = pkgdir(DataAssimilationBenchmarks) * "/src/data/" * method * "/"where the file name is written dynamically according to the selected parameters as follows:
method * "_" * model *
"_state_seed_" * lpad(seed, 4, "0") *
"_diff_" * rpad(diffusion, 5, "0") *
"_sysD_" * lpad(sys_dim, 2, "0") *
"_obsD_" * lpad(obs_dim, 2, "0") *
"_obsU_" * rpad(obs_un, 4, "0") *
"_gamma_" * lpad(γ, 5, "0") *
"_nanl_" * lpad(nanl, 5, "0") *
"_tanl_" * rpad(tanl, 4, "0") *
"_h_" * rpad(h, 4, "0") *
"_lag_" * lpad(lag, 3, "0") *
"_shift_" * lpad(shift, 3, "0") *
"_mda_" * string(mda) *
"_nens_" * lpad(N_ens, 3,"0") *
"_stateInfl_" * rpad(round(s_infl, digits=2), 4, "0") *
".jld2"DataAssimilationBenchmarks.SmootherExps.single_iteration_ensemble_param — Methodsingle_iteration_ensemble_param((time_series::String, method::String, seed:Int64,
nanl::Int64, lag::Int64, shift::Int64, mda::Bool,
obs_un::Float64, obs_dim::Int64, γ::Float64,
p_err::Float64, p_wlk::Float64, N_ens::Int64,
s_infl::Float64, p_infl::Float64)::NamedTuple)SIEnKS joint state-parameter estimation twin experiment.
Output from the experiment is saved in a dictionary of the form,
data = Dict{String,Any}(
"fore_rmse" => fore_rmse,
"filt_rmse" => filt_rmse,
"post_rmse" => post_rmse,
"param_rmse" => para_rmse,
"fore_spread" => fore_spread,
"filt_spread" => filt_spread,
"post_spread" => post_spread,
"param_spread" => para_spread,
"method" => method,
"seed" => seed,
"diffusion" => diffusion,
"dx_params" => dx_params,
"param_truth" => param_truth,
"sys_dim" => sys_dim,
"obs_dim" => obs_dim,
"obs_un" => obs_un,
"γ" => γ,
"p_wlk" => p_wlk,
"p_infl" => p_infl,
"nanl" => nanl,
"tanl" => tanl,
"lag" => lag,
"shift" => shift,
"mda" => mda,
"h" => h,
"N_ens" => N_ens,
"s_infl" => round(s_infl, digits=2),
"p_infl" => round(p_infl, digits=2)
)
if haskey(ts, "diff_mat")
data["diff_mat"] = ts["diff_mat"]
endExperiment output is written to a directory defined by
path = pkgdir(DataAssimilationBenchmarks) * "/src/data/" * method * "-single-iteration/"where the file name is written dynamically according to the selected parameters as follows:
method * "-single-iteration_" * model *
"_param_seed_" * lpad(seed, 4, "0") *
"_diff_" * rpad(diffusion, 5, "0") *
"_sysD_" * lpad(sys_dim, 2, "0") *
"_obsD_" * lpad(obs_dim, 2, "0") *
"_obsU_" * rpad(obs_un, 4, "0") *
"_gamma_" * lpad(γ, 5, "0") *
"_paramE_" * rpad(p_err, 4, "0") *
"_paramW_" * rpad(p_wlk, 6, "0") *
"_nanl_" * lpad(nanl, 5, "0") *
"_tanl_" * rpad(tanl, 4, "0") *
"_h_" * rpad(h, 4, "0") *
"_lag_" * lpad(lag, 3, "0") *
"_shift_" * lpad(shift, 3, "0") *
"_mda_" * string(mda) *
"_nens_" * lpad(N_ens, 3,"0") *
"_stateInfl_" * rpad(round(s_infl, digits=2), 4, "0") *
"_paramInfl_" * rpad(round(p_infl, digits=2), 4, "0") *
".jld2"DataAssimilationBenchmarks.SmootherExps.single_iteration_ensemble_state — Methodsingle_iteration_ensemble_state((time_series::String, method::String, seed::Int64,
nanl::Int64, lag::Int64, shift::Int64, mda::Bool,
obs_un::Float64, obs_dim::Int64, γ::Float64,
N_ens::Int64, s_infl::Float64})::NamedTuple)SIEnKS state estimation twin experiment.
Output from the experiment is saved in a dictionary of the form,
data = Dict{String,Any}(
"fore_rmse" => fore_rmse,
"filt_rmse" => filt_rmse,
"post_rmse" => post_rmse,
"fore_spread" => fore_spread,
"filt_spread" => filt_spread,
"post_spread" => post_spread,
"method" => method,
"seed" => seed,
"diffusion" => diffusion,
"dx_params" => dx_params,
"sys_dim" => sys_dim,
"obs_dim" => obs_dim,
"obs_un" => obs_un,
"γ" => γ,
"nanl" => nanl,
"tanl" => tanl,
"lag" => lag,
"shift" => shift,
"mda" => mda,
"h" => h,
"N_ens" => N_ens,
"s_infl" => round(s_infl, digits=2)
)
if haskey(ts, "diff_mat")
data["diff_mat"] = ts["diff_mat"]
endExperiment output is written to a directory defined by
path = pkgdir(DataAssimilationBenchmarks) * "/src/data/" * method * "-single-iteration/"where the file name is written dynamically according to the selected parameters as follows:
method * "-single-iteration_" * model *
"_state_seed_" * lpad(seed, 4, "0") *
"_diff_" * rpad(diffusion, 5, "0") *
"_sysD_" * lpad(sys_dim, 2, "0") *
"_obsD_" * lpad(obs_dim, 2, "0") *
"_obsU_" * rpad(obs_un, 4, "0") *
"_gamma_" * lpad(γ, 5, "0") *
"_nanl_" * lpad(nanl, 5, "0") *
"_tanl_" * rpad(tanl, 4, "0") *
"_h_" * rpad(h, 4, "0") *
"_lag_" * lpad(lag, 3, "0") *
"_shift_" * lpad(shift, 3, "0") *
"_mda_" * string(mda) *
"_nens_" * lpad(N_ens, 3,"0") *
"_stateInfl_" * rpad(round(s_infl, digits=2), 4, "0") *
".jld2"